Input Data
Source
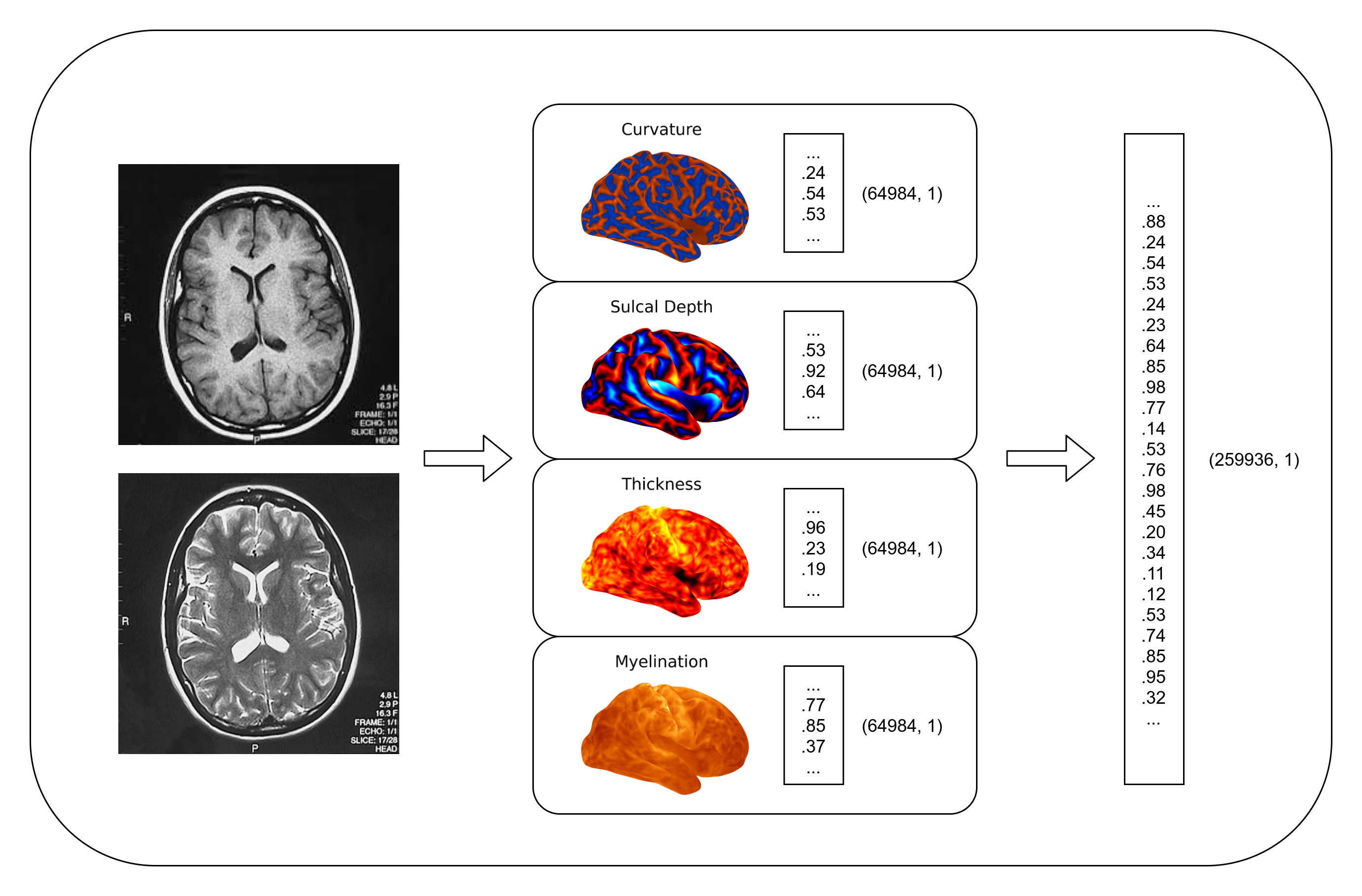
Data is sourced from the baseline (9-10 yr old) ABCD Study release NDA Collection 3165 Release 1.1.0. Data used within this study are the sMRI outputs of a modified HCP style pipeline. We downloaded for each available subject their left and right hemisphere curvature, sulcal depth, cortical thickness and unsmoothed myelin map, each in the standard FS LR 32k vertex space. Likewise, we additionally downloaded each subject’s automatically computed FreeSurfer ROI stats files.
Modality
We chose to use structural MRI surfaces. That said, the idea of parcellations easily extends to task-based fMRI and resting state fMRI just as easily as dMRI. The choice to use structural MRI surfaces was therefore somewhat arbitrary, but given its ubiquity and the amount of available studies which employ it, it may not be a bad choice. Future work may very well consider different modalities or explicitly multi-modal fusion.
Sample Size
One of the big strengths of this study is that we use data from 9,432 participants! This allows us to perform ML and be more confident about our results than smaller studies would allow. Importantly, studies with small sample sizes are well known to produce spurious ML results with large CIs.
Downloading
In order to download the used data, we employed the following download tool: NDA ABCD Downloader. The following subjects of data were specified when downloading for all available subjects:
- derivatives.anat.space-fsLR32k_curv
- derivatives.anat.space-fsLR32k_desc-smoothed_myelinmap
- derivatives.anat.space-fsLR32k_myelinmap
- derivatives.anat.space-fsLR32k_sulc
- derivatives.anat.space-fsLR32k_thickness
- derivatives.anat.stats
The data is then moved to the raw/ folder upon successful download. Unfortunately, due to data privacy issues, this data cannot be shared directly. If interested in working with ABCD Study data, please find more information on their website https://abcdstudy.org/.