Resampling Parcellations
For this project we gathered a large collection of different Existing Parcellations, which often required various resampling procedures depending on their native space and type (e.g., ‘Hard’ vs. ‘Soft’). The different procedures decided upon are listed below. That said, some parcellation were already in this space and only needed minimal pre-processing (e.g., converting from saved matlab arrays, or concatenating lh and rh files saved separate).
Resampling from FreeSurfer Standard Space
From fsaverage surface space, resampling to fs LR 32k space was conducted with tools available from the Human Connectome Project Workbench (Marcus, 2011). See wrapper function surf_to_surf. This command has alternate versions with flag ‘-label-resample’ for resampling static / hard parcellations and ‘-metric-resample’ for resampling each parcel of a probabilistic parcellation.
Once in fs LR 32k space we further masked out the medial wall for all parcellations according to the defined medial wall in fs LR 32k space.
Resampling from Volumetric Standard Space
The Brainnetome parcellation is used here as an example of a static parcellation originally in volumetric space that must be resampled to fs LR 32k surface space.
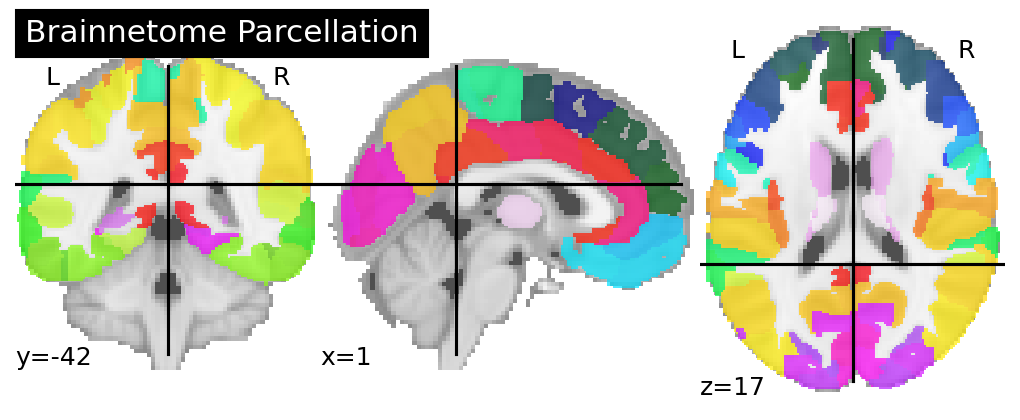
To re-sample static / “hard” volumetric parcellations, we first converted them to “soft” parcellations, in practice treating each region of interest as a separate volume (where that region is labelled with 1 and everywhere else 0).
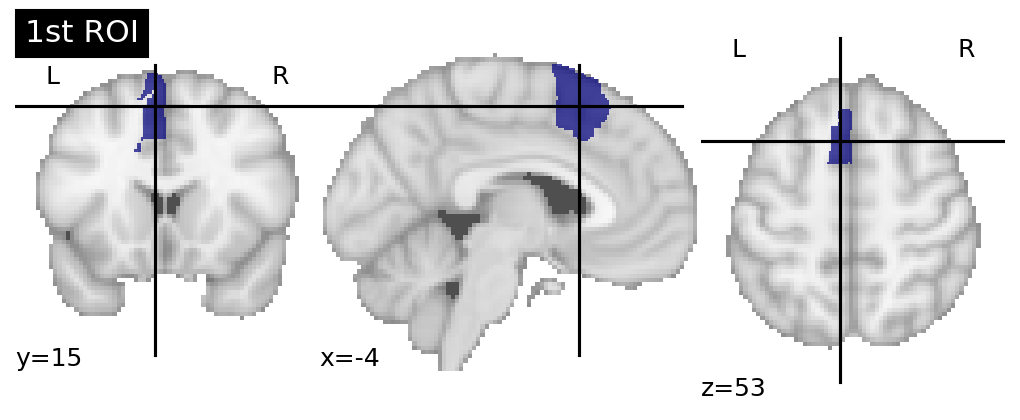
These separate volumes are then individually resampled using Registration Fusion a technique from Wu et al. 2018. The result of this resampling is that ROI as projected to fsaverage space.
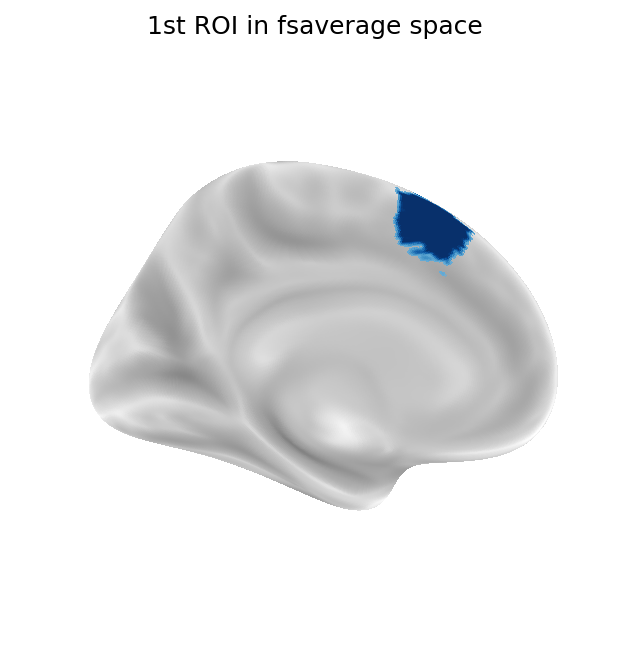
Next, each ROI is resampled from fsaverage space to fs LR 32k space in the same manner as with surfaces originally in fsaverage space.
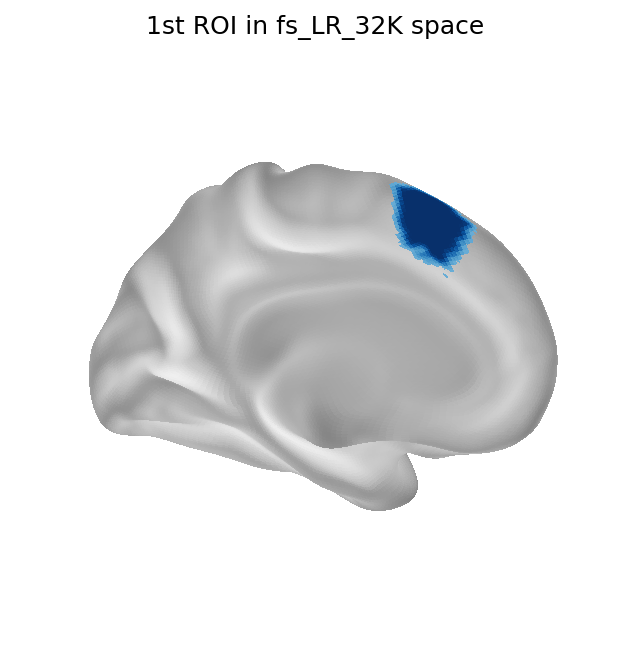
The above process is repeated for all ROIs separately, then in order to re-combine them, a simple argmax operation is carried out across all separate ROI’s surfaces, where each vertex is assigned to the ROI with the highest value.
Lastly, the medial wall is masked out, according to the medial mall defined by FS LR 32k space.

Code
The ‘meat’ of the exact code used to re-sample parcellations can be found at setup/helpers.py which includes the source code for functions called to process specific parcellations in script setup/process_parcs.py. The functions contained here are for the most part python wrappers around different command line tools. For example:
def conv_to_gifti(lh_loc, rh_loc):
temp_name = 'temp'
setup_fs_loc = os.path.join(freesurfer_loc, 'SetUpFreeSurfer.sh')
fs_cmd = 'source ' + setup_fs_loc
lh_gifti = 'L.' + temp_name + '.gii'
rh_gifti = 'R.' + temp_name + '.gii'
# Write full command to temp file and run
with open('temp.sh', 'w') as f:
f.write(fs_cmd + ' && ')
f.write('mri_convert ' + lh_loc + ' ' + lh_gifti + ' && ')
f.write('mri_convert ' + rh_loc + ' ' + rh_gifti)
os.system('bash temp.sh')
os.remove('temp.sh')
os.remove(lh_loc)
os.remove(rh_loc)
return lh_gifti, rh_gifti
Which is used to convert surface files from freesurfer format to gifti via the freesurfer command mri_convert.
Libraries / External Code Used
The re-sampling required the use of a number of external libraries including: