Parcellations
Within this project we consider two main sources for surface parcellations, Existing and Random. We also considered two additional sources: Downsampled / Icosahedron and FreeSurfer Extracted.
Breakdown by type:
- Random: 130
- Existing: 82 (Static: 68 - Prob: 14)
- Icosahedron: 6
- FreeSurfer: 2
- Total: 220
Existing Parcellations
In total, we assessed 82 existing parcellations. Parcellations available at multiple scales were assessed at every scale; in some cases where multiple versions of the same parcellation were available (e.g., from different re-sampling procedures or with different post processing applied), both versions were tested. 68 of the 82 parcellations were static or “hard” parcellations, in which each vertex is labelled as a part of exactly one parcel. We additionally considered 14 probabilistic or “soft” parcellations, where each parcel is represented by a set of probabilities or weightings across the whole surface or volume.
As we were interested in having the parcellations match the space of the data, all parcellations if not already is fs LR 32k space were re-sampled accordingly. For a detailed look at how resampling parcellations between different spaces see resampling.
The existing parcellations used are listed below:
| Name | # of Parcellations | Type | Reference |
| Schaefer Local-global | 10 (scales 100-1000) | Hard | Schaefer 2018 |
| Gordon | 3 (different sources) | Hard | Gordon 2016 |
| Brodmann Areas | 1 | Hard | Brodmann 1909 |
| VDG11b | 1 | Hard | Van Essen 2012 |
| HCP-MMP | 3 (different sources) | Hard | Glasser 2016 |
| Automatic Anatomical Labeling (AAL) | 2 (different sources) | Hard | Tzourio-Mazoyer 2002 |
| Baldassano | 1 | Hard | Baldassano 2015 |
| Desikan | 2 (different sources) | Hard | Desikan 2006 |
| Destrieux | 2 (different sources) | Hard | Destrieux 2010 |
| Brainnetome | 2 (different sources) | Hard | Fan 2016 |
| Power | 2 (different sources) | Hard | Power 2011 |
| Shen 268 Parcels | 2 (different sources) | Hard | Shen 2013 |
| Shen 368 Parcels | 1 | Hard | Salehi 2020 |
| Yeo | 3 (7 Networks, 17 Networks and parcel level version) | Hard | Yeo 2014 |
| DiFuMo | 5 (scales 64-1028) | Soft | Dadi 2020 |
| MIST | 9 (scales 7-444) | Hard | Urchs 2019 |
| AICHA | 1 | Hard | Joliot 2015 |
| Economo | 1 | Hard | von Economo 2915 |
| NSPN500 | 1 | Hard | Whitaker 2016 |
| Oasis | 1 | Hard | Sabuncu 2011 |
| SJH | 1 | Hard | Harrison 2015 |
| Allen | 1 | Soft | Allen 2011 |
| BASC | 9 (scales 9-444) | Hard | Bellec 2013 |
| MSDL | 1 | Soft | Varoquaux 2011 |
| Harvard Oxford | 4 (different versions) | Hard / Soft | Jenkinson 2012 |
| Craddock | 4 (different versions) | Soft | Craddock 2012 |
| Smith ICA | 2 (different versions) | Soft | Smith 2009 |
| CPAC | 1 | Hard | Craddock 2013 |
| Hammersmith | 1 | Hard | Hammers 2003 |
| JuBrain | 1 | Hard | Eickhoff 2005 |
| MICCAI | 1 | Hard | 2012 MICCAI Challenge_Data |
| Slab | 2 (907 and 1068) | Hard | Sripada 2014 |
| Princeton Visual | 1 | Hard | Wang 2015 |
Click the parcellation name link to see that parcellation plotted
See also the folder raw/ which contains the ‘raw’ existing parcellations, before any preprocessing or re-sampling conducted by this project, also included are information on how they can be downloaded. See also the script setup/process_parcs.py which includes the specific code used to process the data from the raw/ folder into the Final Parcellations Used.
Random Parcellations
This project uses the idea of random surface parcellations extensively. We generated 5 random parcellations for of the following sizes in the base experiment:
10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 150, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1500, 2000, 3000, 4000, 5000, 6000
Click a parcellation size to see those random parcellations
Random parcellations are generated as follows: For a random parcellation of size N, N random points are first selected at random across both hemisphere’s 59,412 vertices (medial wall vertices excluded). Each selected point is then assigned as the seed of a new region and is randomly assigned a size probability between 0 and 1. Next, a region is randomly selected according to a weighted random choice between all regions (e.g., if a region was assigned an initial probability of .5 it would be picked on average twice as often as a region assigned .25). A random vertex is then added to the selected region from the list of valid neighboring unassigned vertices. This sequence, of selecting a region and adding one valid vertex, is repeated until all regions have no unassigned neighbors and therefore all non-medial wall vertices are assigned to a region.
Example of generating a random parcellation:
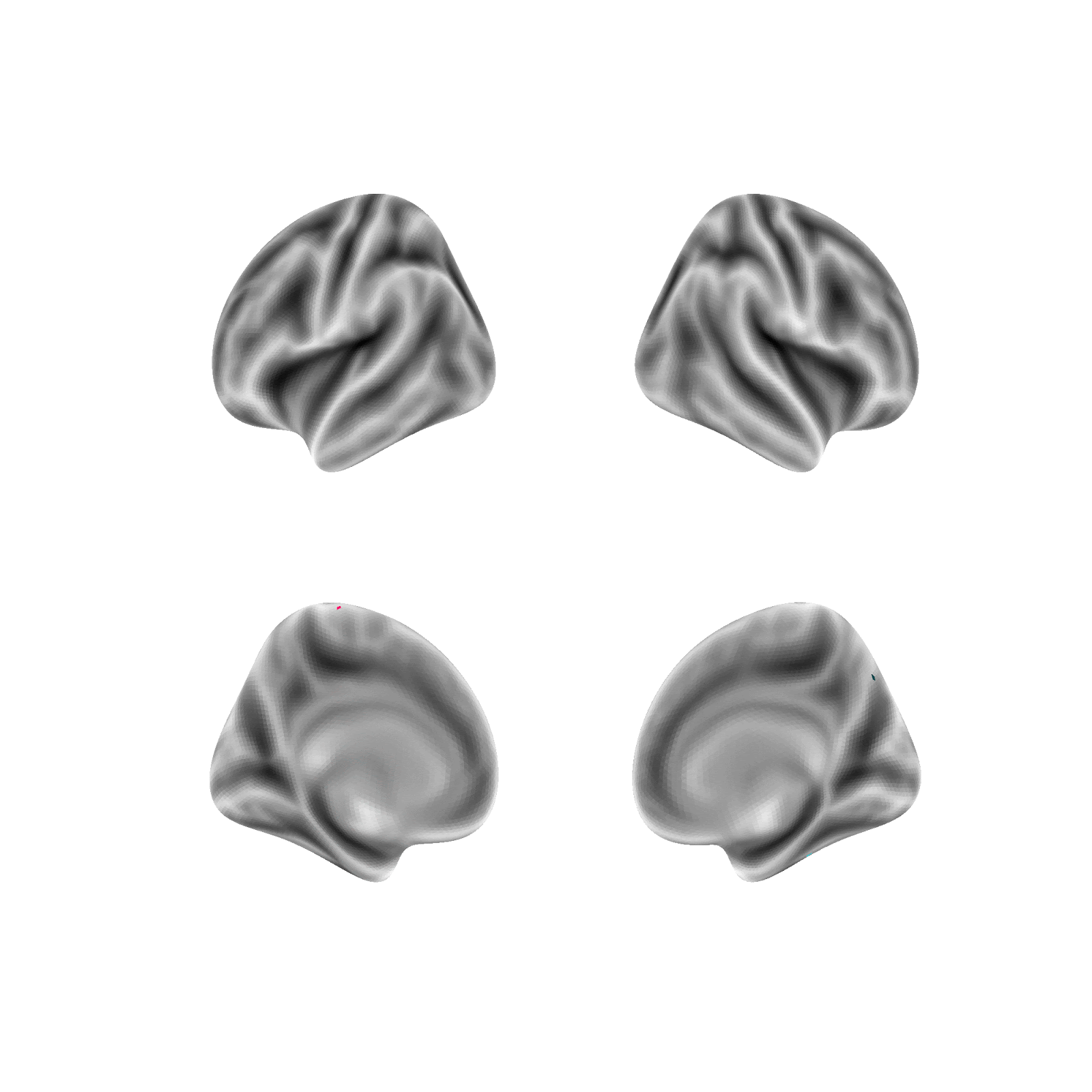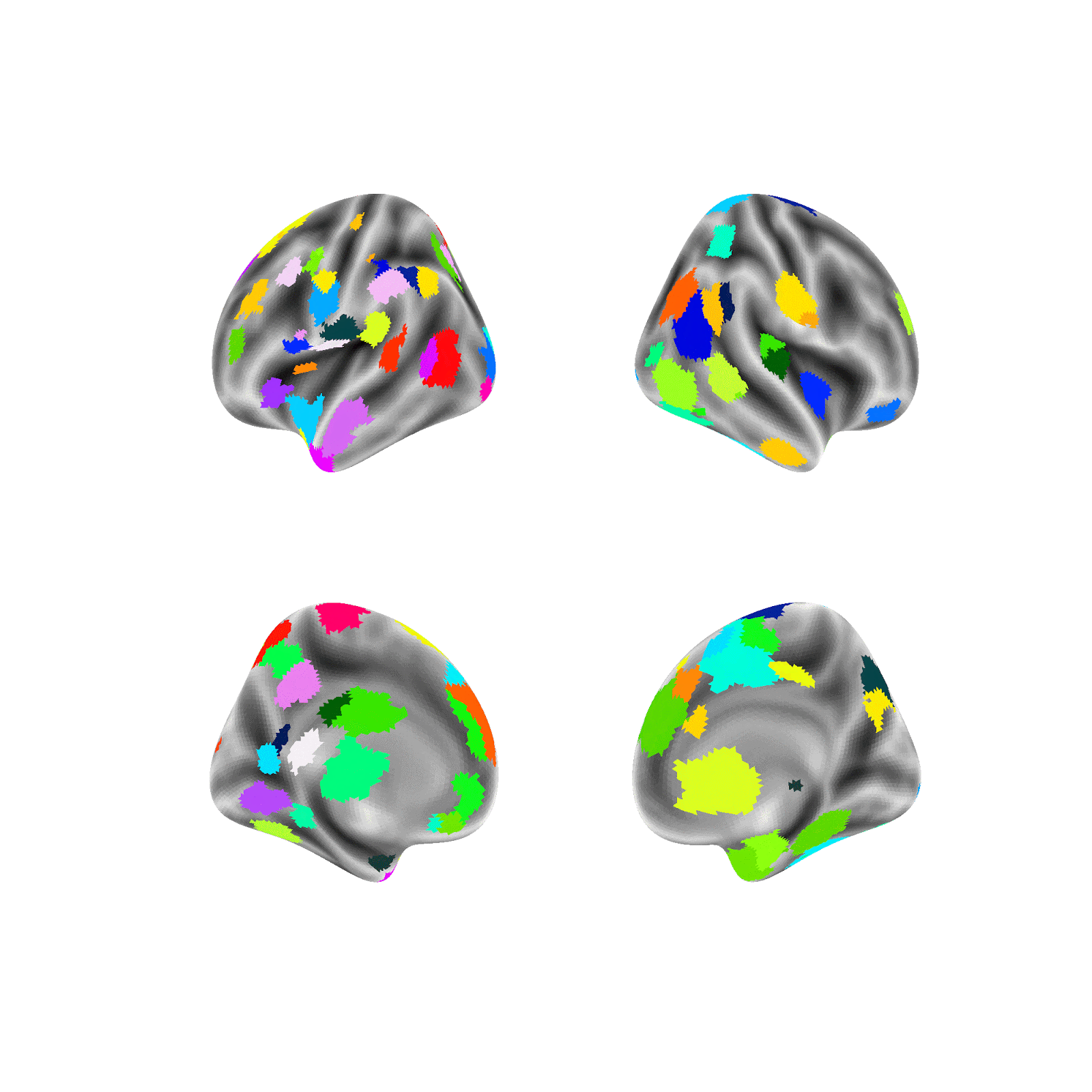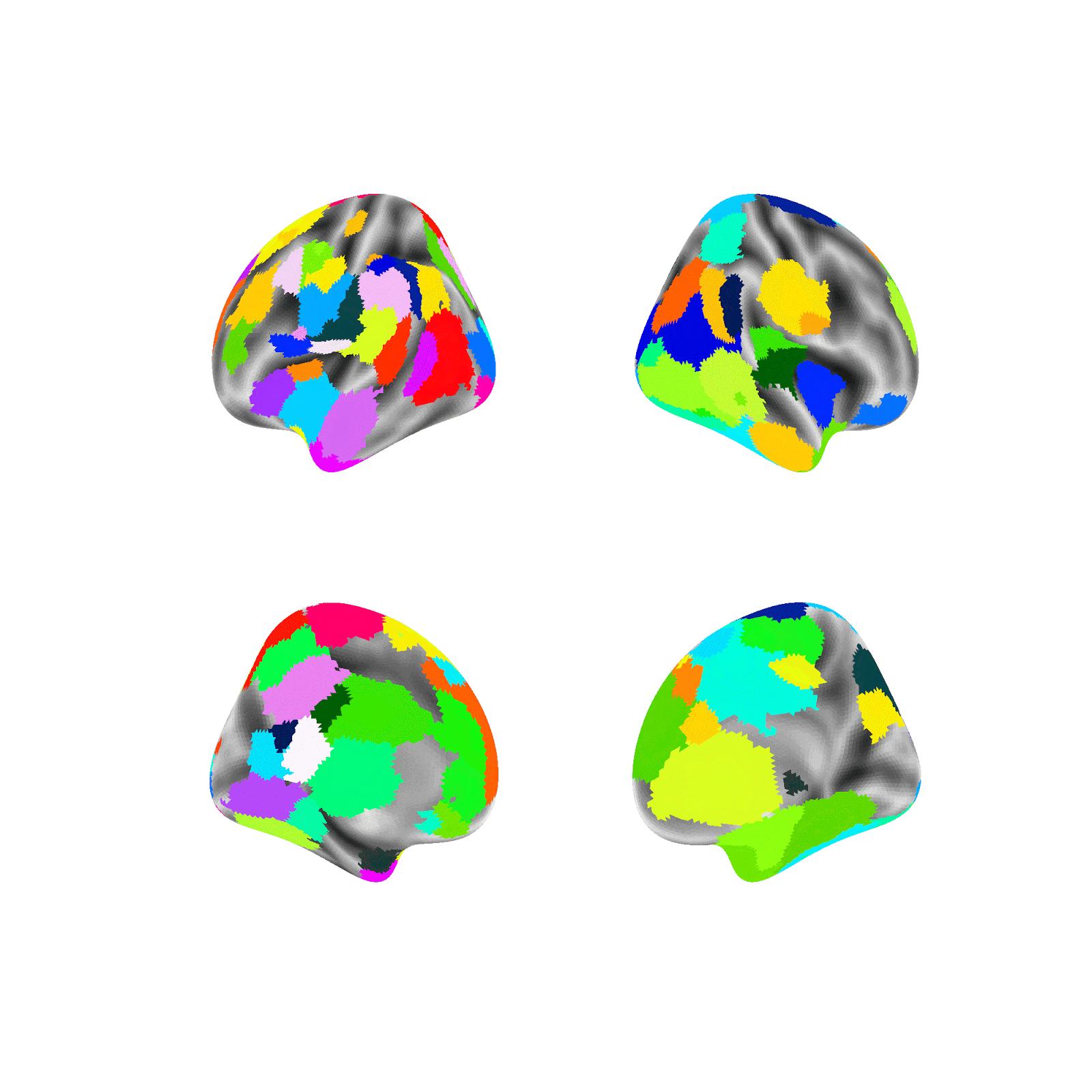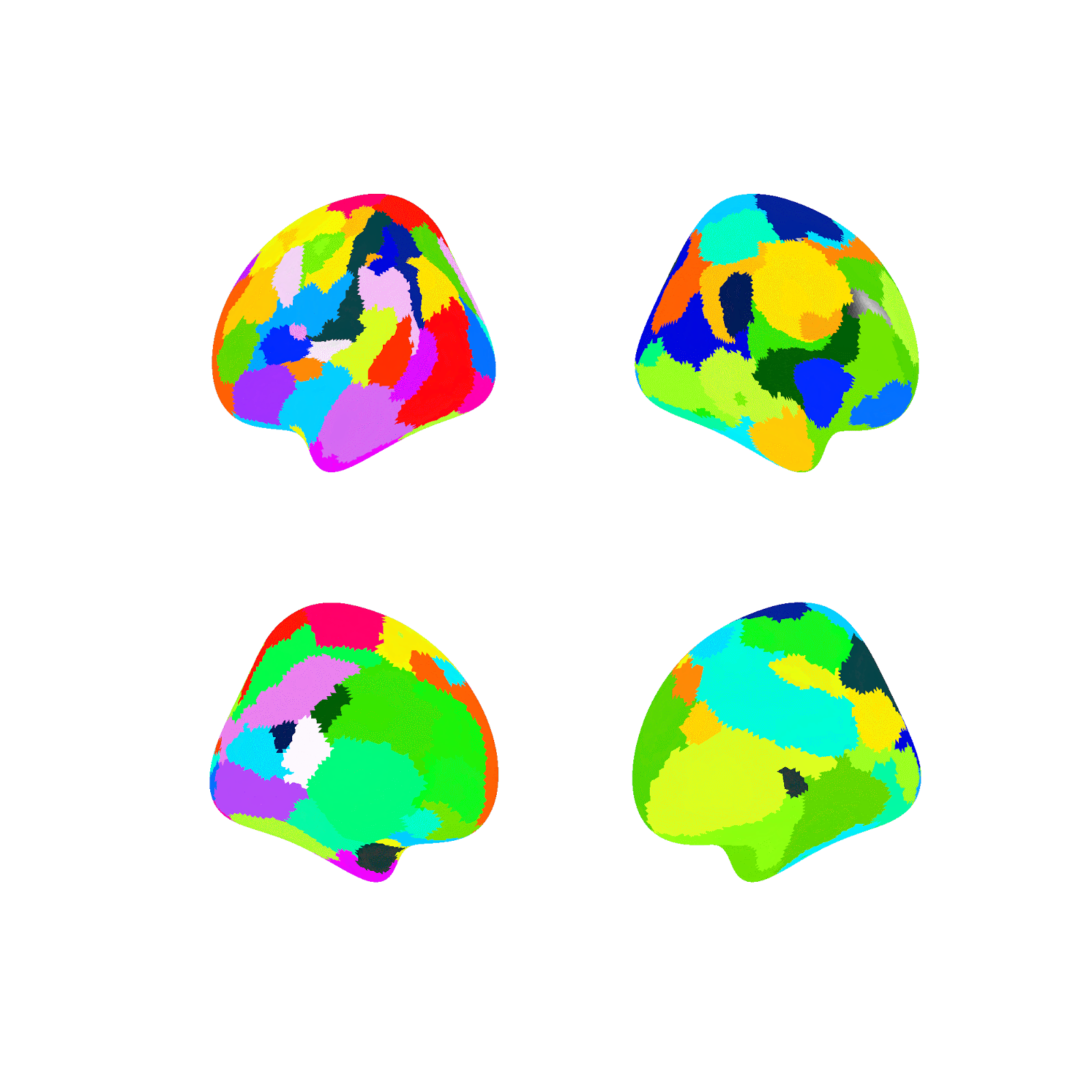
Note: The above example, in contrast to the random parcellations generated in this project, is in fsaverage5 space (vs. fs_LR_32k) and doesn’t mask the medial wall (the medial wall is masked in this project).
Source code for generating random parcellations is implemented and available through the Brain Predictability toolbox (BPt), specifically here.
Random parcellations within this project are generated in the setup/process_random_parcels.py script.
Icosahedron Parcellations
We test 6 different downsampled icosahedron parcellations (with medial wall removed). These spanned sizes: 42, 162, 362, 642, 1002, 1442. See parcellations plotted.
FreeSurfer ROIs
The last parcellation we tested was actually using the FreeSurfer extracted ROIs directly. These differ from the other tested parcellations both in how values are generated (FreeSurfer extracts values in an individual’s native space whereas we extract values from data warped to a common space) in addition to the surface modalities used (only average thickness, surface area and mean curvature are employed, which differs from the features used in the base analyses). These were extracted in order to provide a comparison with with other common a ML neuroimaging analyses.
The code for generating the dataset of FreeSurfer ROIs is found here.
Source
To access the final saved numpy array representations of the parcellations used, see Final Parcellations Used. These are saved as numpy arrays corresponding the fs_LR_32K space, in the case of probabilistic parcellation are saved with shape (64984, number of parcels), otherwise are saved as a flat array of 64984 vertex (left hemi first).
These parcellations are also all visualized here.
References
-
Arslan, S., Ktena, S. I., Makropoulos, A., Robinson, E. C., Rueckert, D., & Parisot, S. (2018). Human brain mapping: A systematic comparison of parcellation methods for the human cerebral cortex.Neuroimage, 170, 5-30.
-
Baldassano, C., Beck, D. M., & Fei-Fei, L. (2015). Parcellating connectivity in spatial maps. PeerJ, 3, e784.
-
Bellec, P. (2013, June). Mining the hierarchy of resting-state brain networks: selection of representative clusters in a multiscale structure. In 2013 International Workshop on Pattern Recognition in Neuroimaging (pp. 54-57). IEEE.
-
Brodmann, K. (1909). Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Barth. Challenges.”NeuroImage, vol. 197, 2019, pp. 652–656., doi:10.1016/j.neuroimage.2018.10.003.
-
Craddock, C., Sikka, S., Cheung, B., Khanuja, R., Ghosh, S. S., Yan, C., … & Milham, M. (2013). Towards automated analysis of connectomes: The configurable pipeline for the analysis of connectomes (c-pac). Front Neuroinform, 42.
-
Craddock, R. C., James, G. A., Holtzheimer III, P. E., Hu, X. P., & Mayberg, H. S. (2012). A whole brain fMRI atlas generated via spatially constrained spectral clustering. Human brain mapping, 33(8), 1914-1928.
-
Dadi, K., Varoquaux, G., Machlouzarides-Shalit, A., Gorgolewski, K. J., Wassermann, D., Thirion, B., and Mensch, A. (2020). Fine-grain atlases of functional modes for fMRI analysis. NeuroImage.
-
Desikan, R. S., Ségonne, F., Fischl, B., Quinn, B. T., Dickerson, B. C., Blacker, D., … & Killiany, R. J. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage, 31(3), 968-980.
-
Destrieux, C., Fischl, B., Dale, A., & Halgren, E. (2010). Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage, 53(1), 1-15.
-
Eickhoff, S. B., Stephan, K. E., Mohlberg, H., Grefkes, C., Fink, G. R., Amunts, K., & Zilles, K. (2005). A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage, 25(4), 1325-1335.
-
Fan, L., Li, H., Zhuo, J., Zhang, Y., Wang, J., Chen, L., … & Jiang, T. (2016). The human brainnetome atlas: a new brain atlas based on connectional architecture. Cerebral cortex, 26(8), 3508-3526.
-
Glasser, M. F., Coalson, T. S., Robinson, E. C., Hacker, C. D., Harwell, J., Yacoub, E., … & Van Essen, D. C. (2016). A multi-modal parcellation of human cerebral cortex. Nature, 536(7615), 171-178.
-
Gordon, E. M., Laumann, T. O., Adeyemo, B., Huckins, J. F., Kelley, W. M., & Petersen, S. E. (2016). Generation and evaluation of a cortical area parcellation from resting-state correlations. Cerebral cortex, 26(1), 288-303.
-
Hammers, A., Allom, R., Koepp, M. J., Free, S. L., Myers, R., Lemieux, L., … & Duncan, J. S. (2003). Three‐dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Human brain mapping, 19(4), 224-247.
-
Harrison, S. J., Woolrich, M. W., Robinson, E. C., Glasser, M. F., Beckmann, C. F., Jenkinson, M., & Smith, S. M. (2015). Large-scale probabilistic functional modes from resting state fMRI. NeuroImage, 109, 217-231.
-
Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W., & Smith, S. M. (2012). Fsl. Neuroimage, 62(2), 782-790.
-
Joliot, M., Jobard, G., Naveau, M., Delcroix, N., Petit, L., Zago, L., … & Tzourio-Mazoyer, N. (2015). AICHA: An atlas of intrinsic connectivity of homotopic areas. Journal of neuroscience methods, 254, 46-59.
-
Power, J. D., Cohen, A. L., Nelson, S. M., Wig, G. S., Barnes, K. A., Church, J. A., … & Petersen, S. E. (2011). Functional network organization of the human brain. Neuron, 72(4), 665-678.
-
Salehi, M., Greene, A. S., Karbasi, A., Shen, X., Scheinost, D., & Constable, R. T. (2020). There is no single functional atlas even for a single individual: Functional parcel definitions change with task. NeuroImage, 208, 116366.
-
Schaefer, A., Kong, R., Gordon, E. M., Laumann, T. O., Zuo, X. N., Holmes, A. J., … & Yeo, B. T. (2018). Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cerebral cortex, 28(9), 3095-3114.
-
Schleicher, A., et al. “Quantitative architectural analysis: a new approach to cortical mapping.” Anatomy and embryology 210.5-6 (2005): 373-386.
-
Shen, X., Tokoglu, F., Papademetris, X., & Constable, R. T. (2013). Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage, 82, 403-415.
-
Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., … & Joliot, M. (2002). Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage, 15(1), 273-289.
-
Urchs, S., Armoza, J., Moreau, C., Benhajali, Y., St-Aubin, J., Orban, P., & Bellec, P. (2019). MIST: A multi-resolution parcellation of functional brain networks. MNI Open Research, 1(3), 3.
-
Van Essen, D. C., Glasser, M. F., Dierker, D. L., Harwell, J., & Coalson, T. (2012). Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cerebral cortex, 22(10), 2241-2262.
-
Varoquaux, G., Gramfort, A., Pedregosa, F., Michel, V., & Thirion, B. (2011, July). Multi-subject dictionary learning to segment an atlas of brain spontaneous activity. In Biennial International Conference on information processing in medical imaging (pp. 562-573). Springer, Berlin, Heidelberg.
-
Wang, L., Mruczek, R. E., Arcaro, M. J., & Kastner, S. (2015). Probabilistic maps of visual topography in human cortex. Cerebral cortex, 25(10), 3911-3931.
-
von Economo, C. F., & Koskinas, G. N. (1925). Die cytoarchitektonik der hirnrinde des erwachsenen menschen. J. Springer.